A project partnership between Scottish Sea Farms, the University of Aberdeen, and BioMar has made important progress in understanding and managing gill disease in farmed Atlantic salmon.
Gill health is vital for fish welfare and can be threatened by changing ocean conditions and disease. Research was conducted across aquaculture sites in Scotland and Tasmania, offering contrasting environments with lower (Scotland) and higher (Tasmania) seawater temperatures. A multidisciplinary approach combining RNA sequencing (RNA-seq), histopathology, microbiota profiling, and high-throughput quantitative PCR (qPCR) validation provided a detailed, multi-layered assessment of gill pathology and its relationship with environmental drivers.
Researchers found that as gill disease worsens, the diversity of beneficial microbes in the gills decreases. They developed a set of over 90 genetic markers, identified through RNA sequencing and validated using qPCR, that may help farmers detect disease earlier and respond more effectively. Many of the validated genes were common to salmon from both Scotland and Tasmania, while others were location-specific.
In parallel, the study identified distinct microbial signatures associated with gill disease progression, with taxa such as Candidatus Branchiomonas, Candidatus Rubidus, and Tenacibaculum consistently linked to pathology.
In Scotland, seasonal variation was a key factor, with gill disease peaking in autumn and correlating strongly with environmental conditions. Microbiome diversity declined significantly after transfer to seawater, at which point Candidatus Clavichlamydia salmonicola became the dominant species. Transcriptomic analysis revealed that the time since seawater transfer was a stronger determinant of gill health than smolt origin, with immune activation and tissue remodeling processes driving much of the observed variation. Non-invasive sampling using RNA extracted from gill swabs was also tested, showing promise for detecting the more abundantly expressed biomarkers.
The evaluation of biomarkers across production sites highlighted a consistent relationship between microbial composition, histopathology, and gene expression patterns. Histological analysis confirmed progressive changes in gill tissue during disease development, while RNA-seq data revealed upregulation of genes involved in immune responses, inflammation, and pathogen defence.
In a subset of fish from the autumn trial, those fed the functional diet showed slightly lower PGD scores than controls, but no statistically significant differences were observed for amoebic gill disease (AGD) scores, PGD scores, or body weight (Fig. 1).
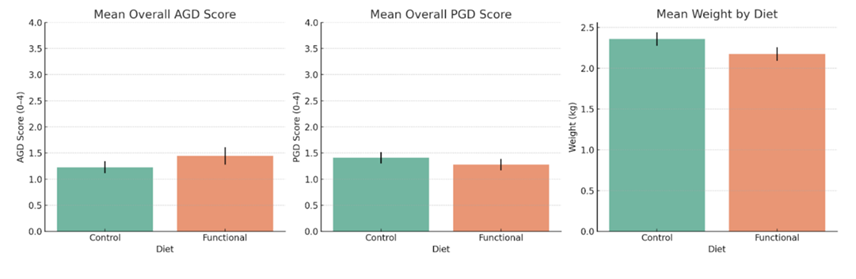
Figure 1. Mean AGD score, PGD score, and body weight of fish fed control or functional diets during the autumn trial. Fish on the functional diet showed slightly lower average PGD scores than controls, but Mann-Whitney U tests indicated no statistically significant differences for AGD (p = 0.27), PGD (p = 0.40), or bodyweight (p = 0.11). Mean body weight was marginally higher in the control group.
Overall, the functional diet did not produce measurable improvements in gill health or growth, likely due to the complexity of gill disease and the additional interventions required during the study, which may have masked any potential dietary effects. These findings suggest that while single nutritional interventions may offer some support, integrated health management approaches are necessary to address gill pathologies effectively.
In Tasmania, biomarker robustness was further demonstrated under higher seawater temperatures. However, functional feeds provided only limited mitigation of gill health impacts in these warmer conditions. Pathogen communities differed notably between regions: while Scottish fish showed signatures of bacterial drivers, Tasmanian fish were heavily affected by Neoparamoeba perurans, the causative agent of AGD. Transcriptomic profiling identified inflammation-related genes as promising early-warning indicators in these outbreaks. Freshwater treatment, although commonly used to manage AGD, produced highly variable outcomes among individual fish, suggesting that more personalised or adaptive management strategies may be required. The findings also underscore the challenges posed by climate change, with rising ocean temperatures likely to intensify the severity of gill disease and reduce the effectiveness of nutritional interventions.
“By enabling earlier disease detection, the outcomes of this project will help support the long-term resilience of the aquaculture sector and strengthen Scotland’s position as a leader in the production of sustainable seafood,” researchers said.
References
Costelloe, EP, Lorgen-Ritchie, M, Król, E, Noguera, P, Bickerdike, R, Tinsley, J, Valdenegro, V, Douglas, A & Martin, SAM. 2025, Microbial and histopathological insights into gill health of Atlantic salmon (Salmo salar) across Scottish aquaculture sites, Aquaculture, vol. 599, 742166. https://doi.org/10.1016/j.aquaculture.2025.742166
Vallarino, MC, Dagen, SL, Costelloe, E, Oyenekan, SI, Tinsley, J, Valdenegro, V, Król, E, Noguera, P & Martin, SAM 2024. Dynamics of Gill Responses to a Natural Infection with Neoparamoeba perurans in Farmed Tasmanian Atlantic Salmon, Animals, vol. 14, no. 16, 2356. https://doi.org/10.3390/ani14162356
Król, E, Noguera, P, Shaw, S, Costelloe, E, Gajardo, K, Valdenegro, V, Bickerdike, R, Douglas, A & Martin, SAM 2020. Integration of Transcriptome, Gross Morphology and Histopathology in the Gill of Sea Farmed Atlantic Salmon (Salmo salar): Lessons from Multi-site Sampling, Frontiers in Genetics, vol. 11, 610. https://doi.org/10.3389/fgene.2020.00610













